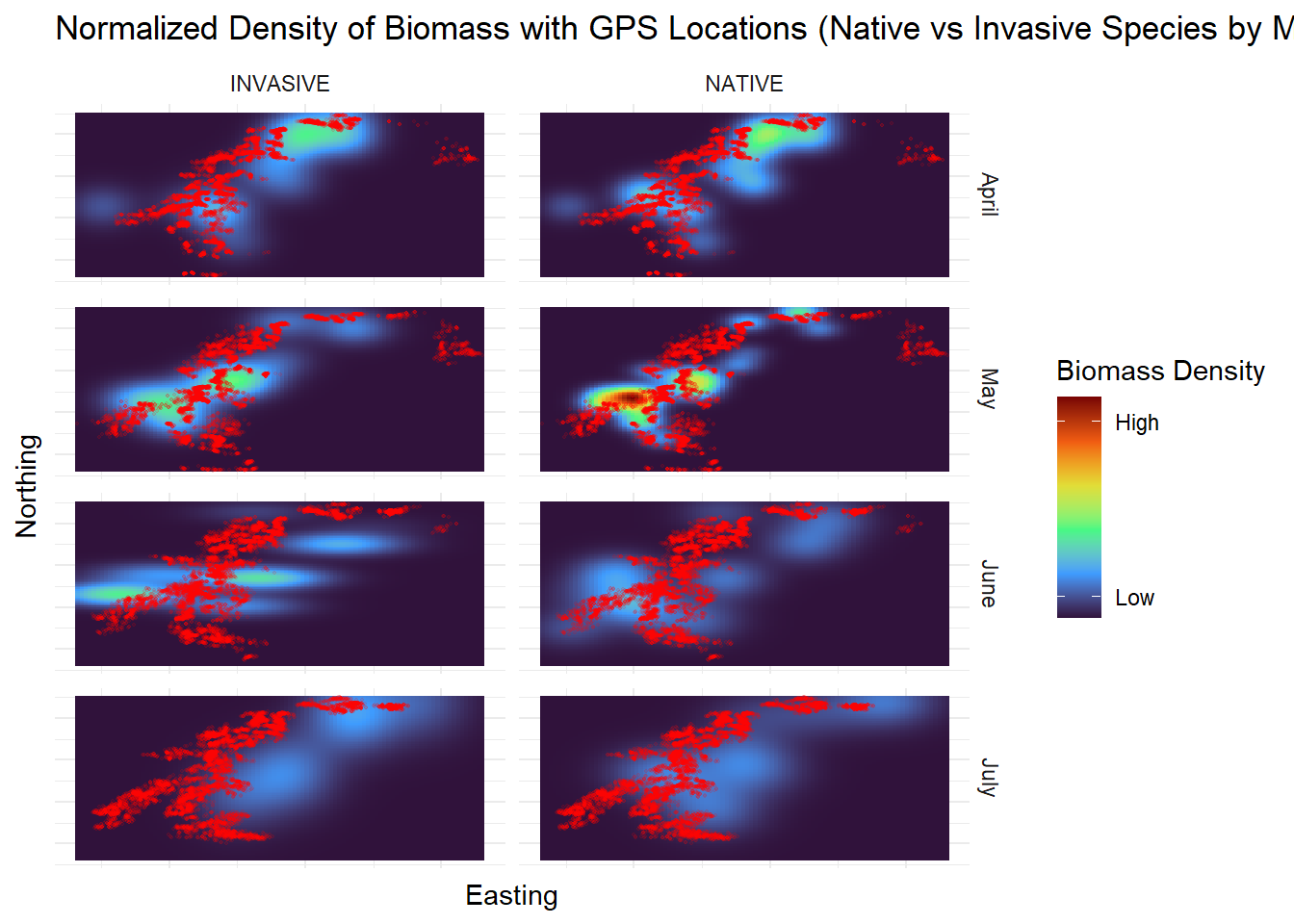
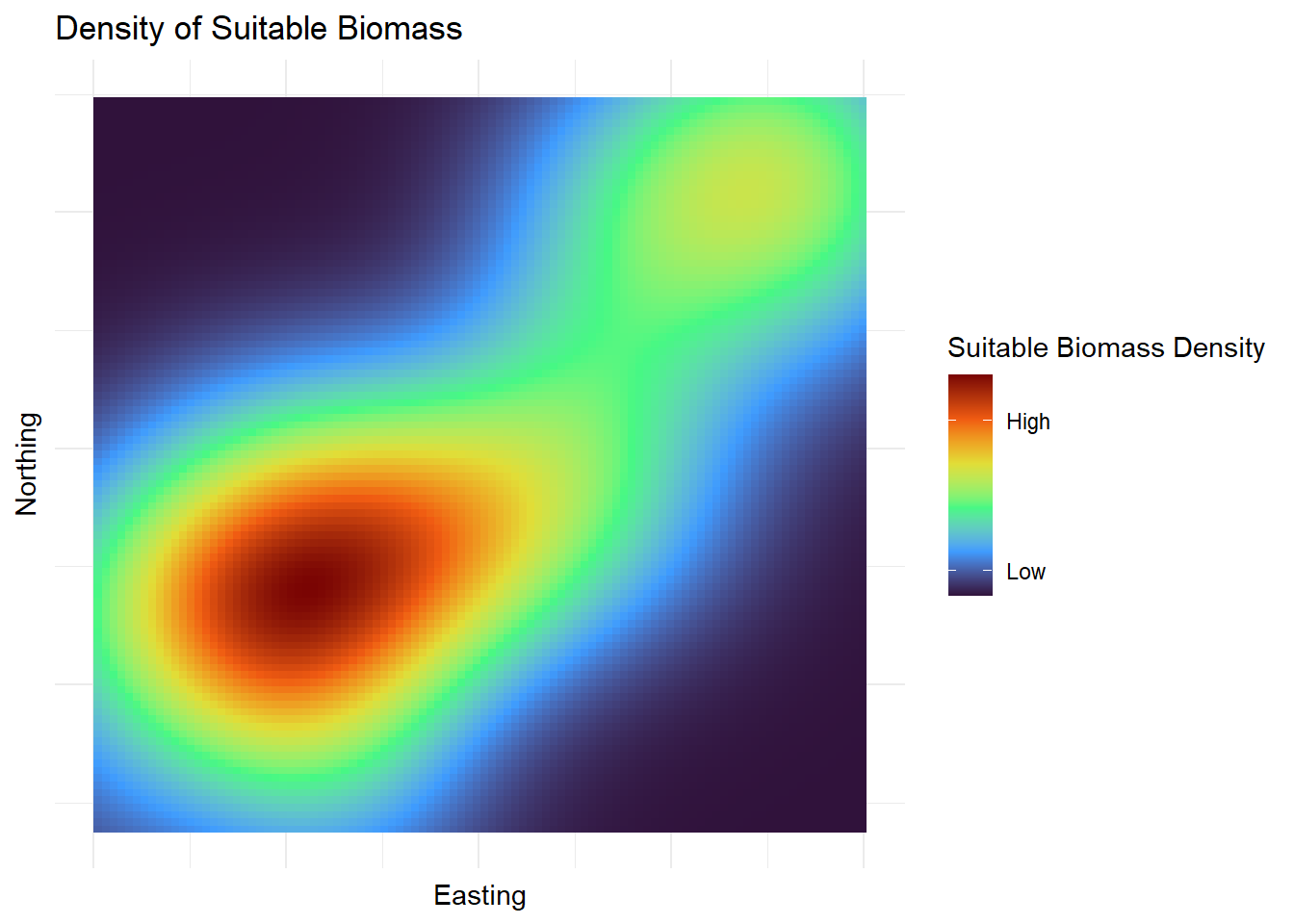
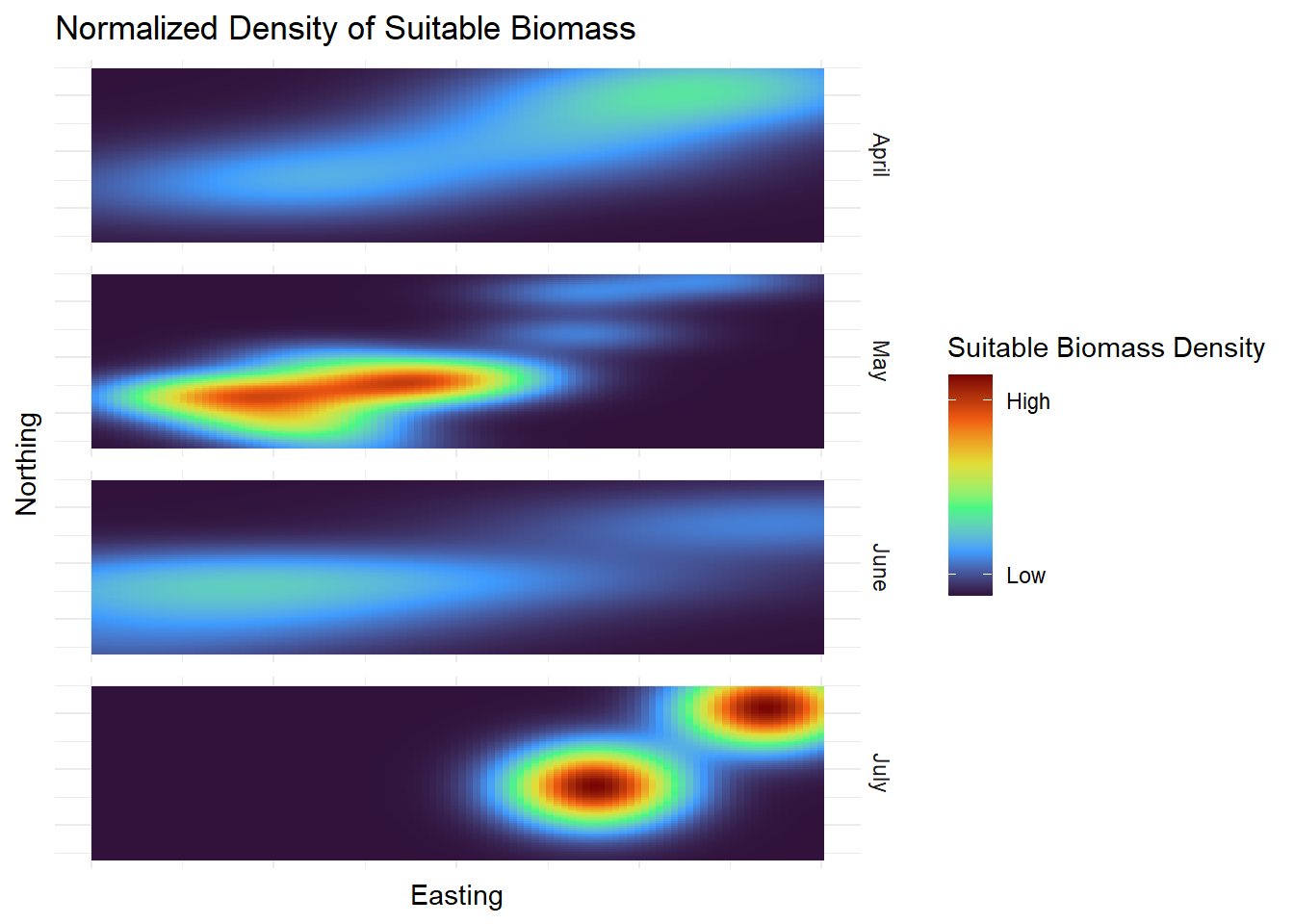
| Biomass |
Quantitative |
This measurement tells us what the weight of dry biomass is for each specific observation |
| TransectID |
Categorical |
This descriptor is a unique ID that tells us which randomized point we sampled |
| Quadrat |
Categorical |
There are 5 quadrats that we sample for each quadrat (20,40,60,80,100) |
| Code |
Categorical |
These are unique codes that describe the family, genius and species for every item observed |
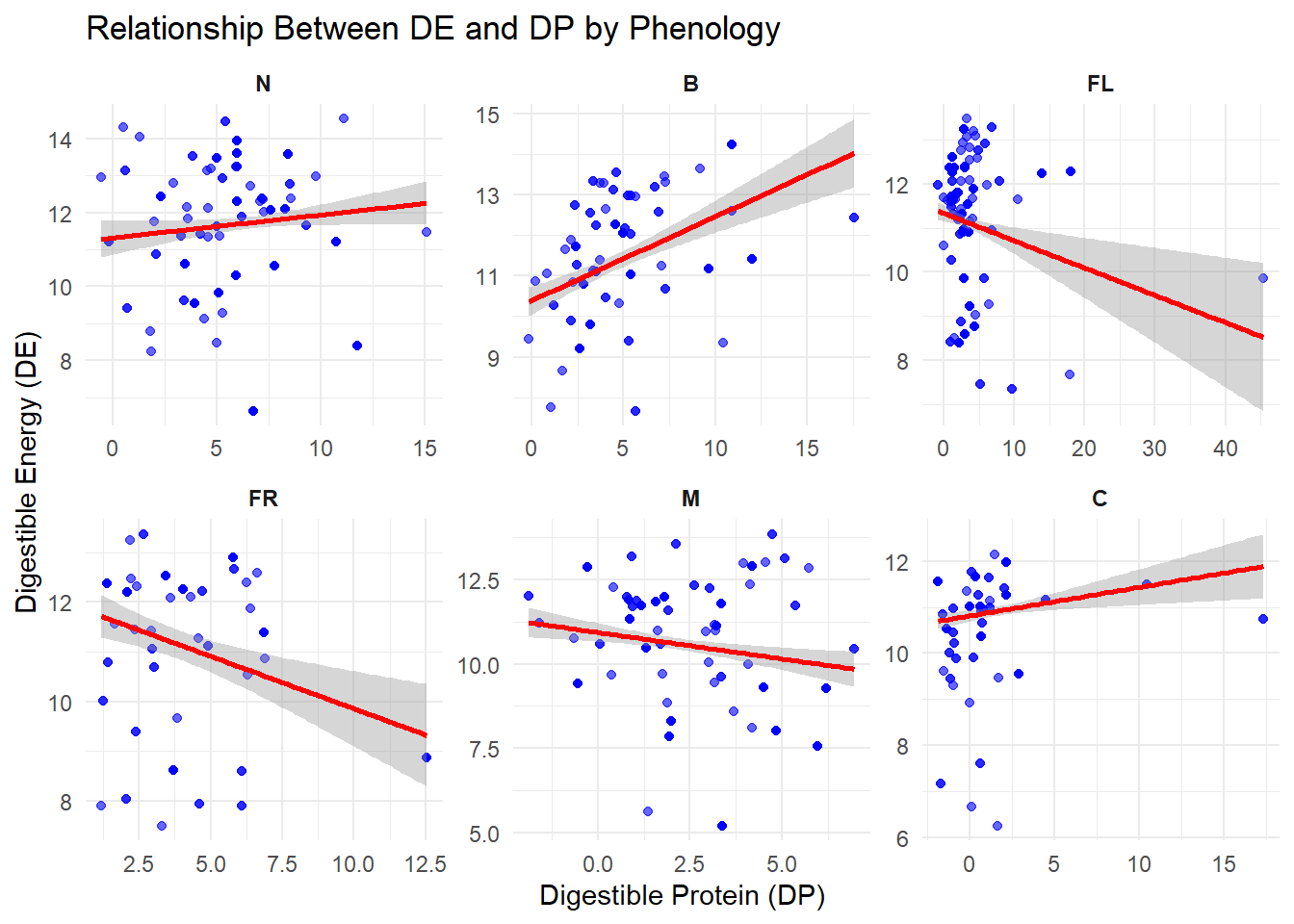
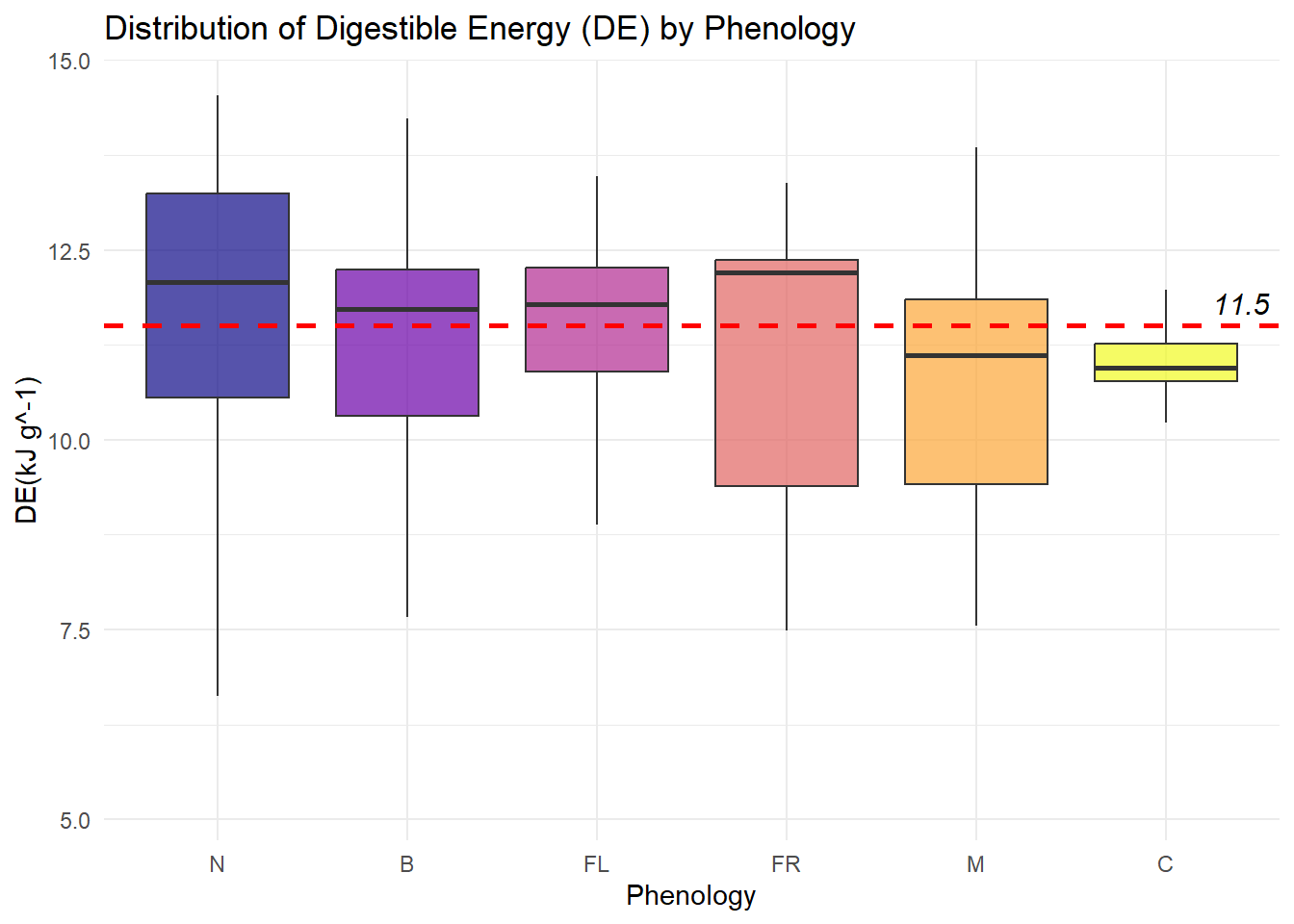
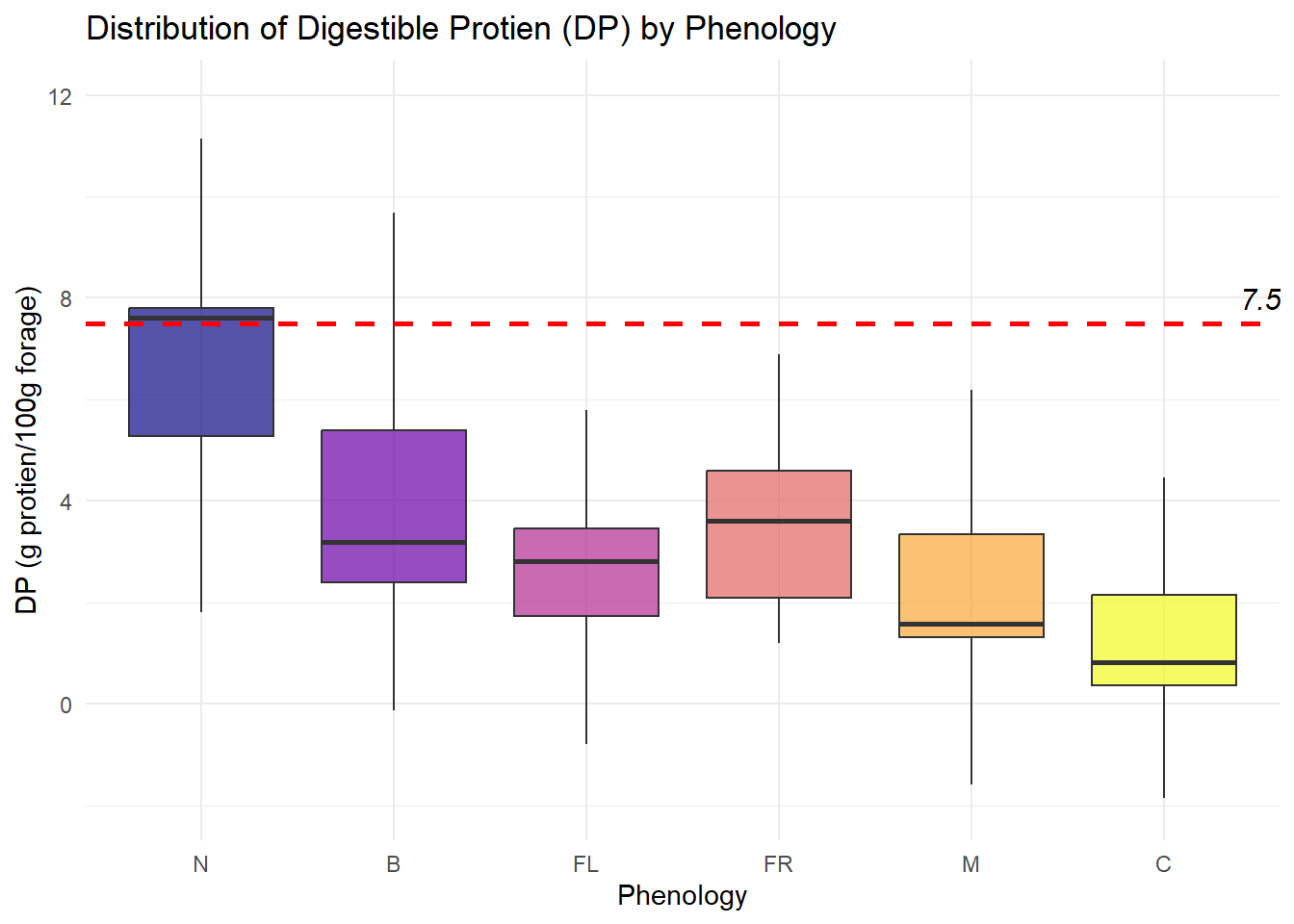
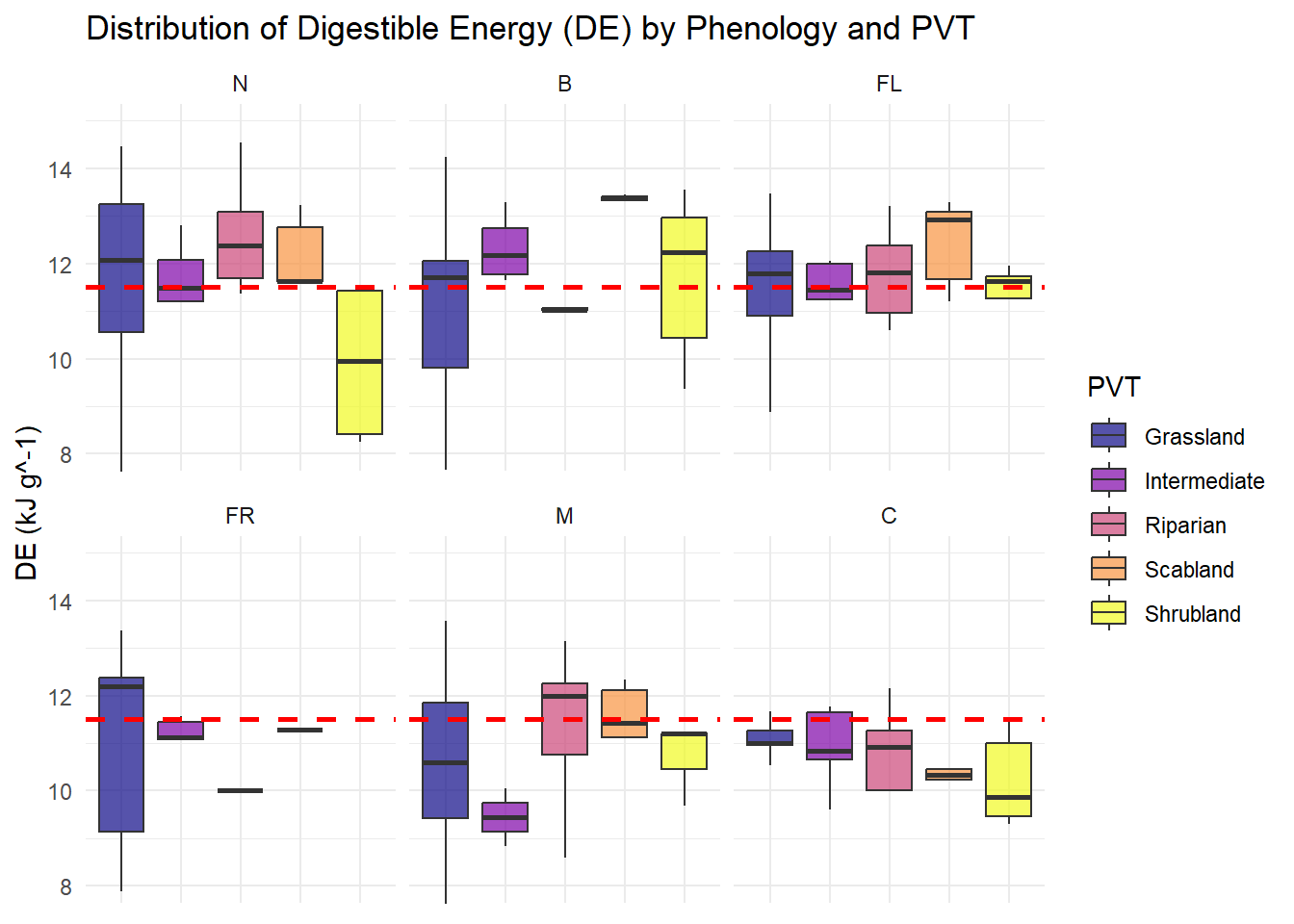
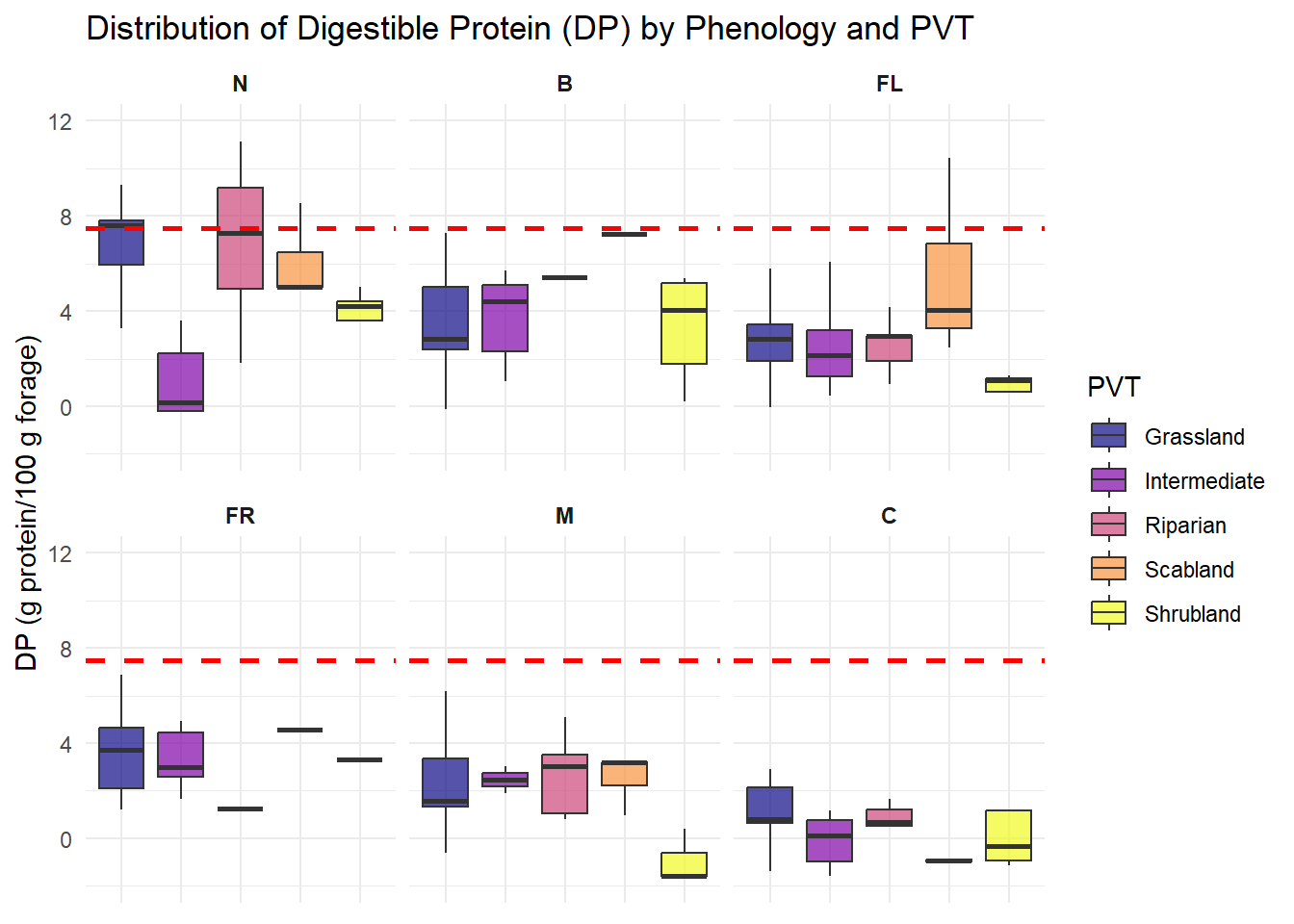
| Phenology |
Categorical |
Each species is assigned a growth stage when we observe it - New, Budding, Flowering, Fruiting, Mature or Cured (N, B, FL, FR, M, C) |
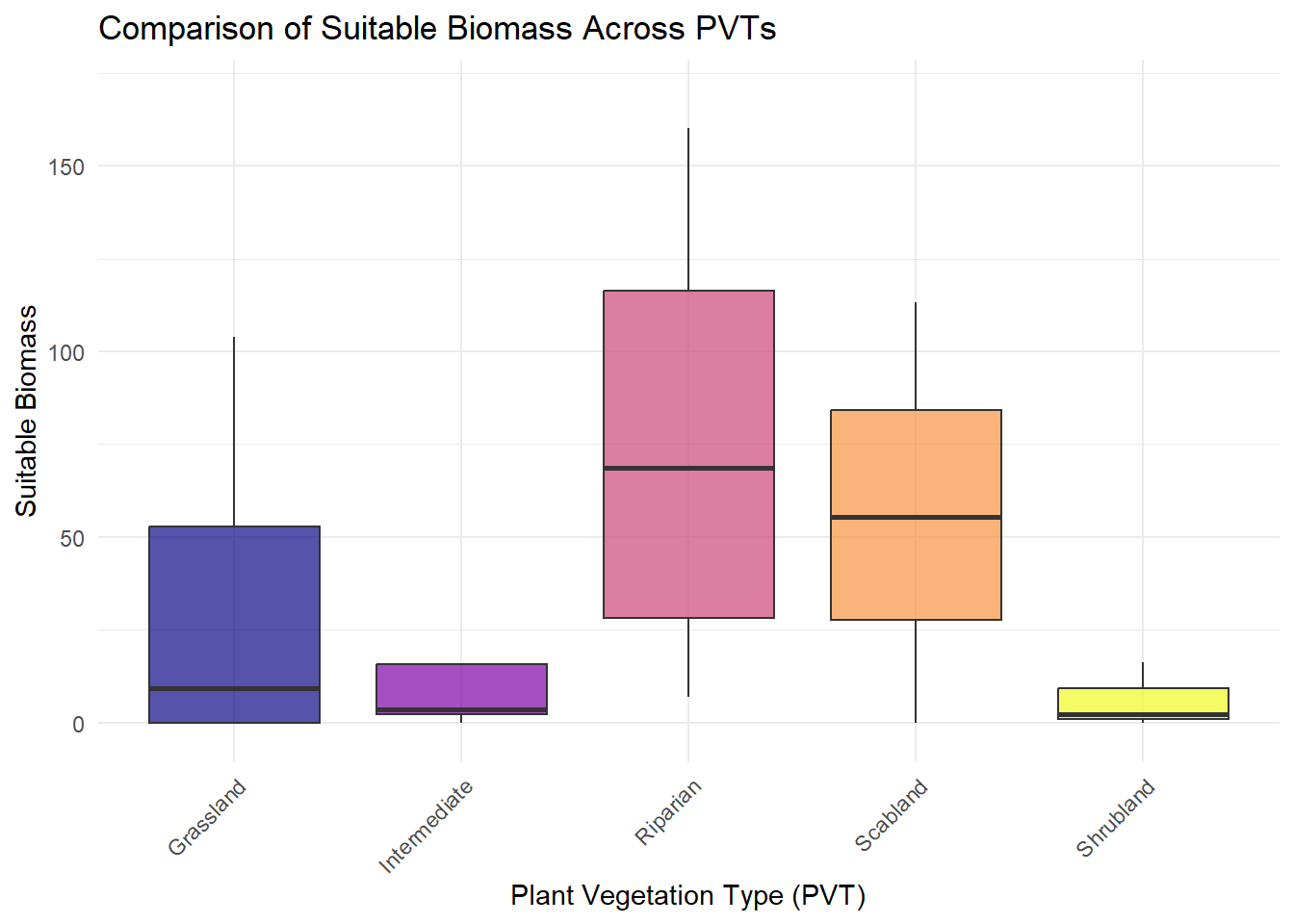
| PVT |
Categorical |
This number describes the vegetation community that was being sampled, we have 5 total for the study area, it is used as part of the descriptor for each plotID |
| percent |
Quantitative |
This measurement tells us the percent cover that each composition_id occupies within a 1x1m quadrat |
| Spp |
Categorical |
This unique ID is slightly more broad and will be used to identify species/phenology combinations within each vegetation type as well as the season |
| Family |
Categorical |
This will be used to group quality data if we do not have enough information to determine the quality down to the smaller scale (genus) |
| Genus |
Categorical |
This will be used to group quality data if we do not have enough information to determine the quality down to the smaller scale (spp) |
| Species |
Categorical |
This will be used to group quality data if we do not have enough information to determine the quality down to the smaller scale (phenological stage) |
| FunctionalGroup |
Categorical |
The smallest grouping variable for my quality data (perennial or annual grass, forb, or shrub) |
| FGNew |
Categorical |
Shortened version of FunctionalGroup |
| CommonName |
Categorical |
This is another identifier for each species, it will likely not be used within the analysis so it could be removed |
| Duration |
Categorical |
This is another category I may use to group quality data based on the growth duration of each species |
| Status |
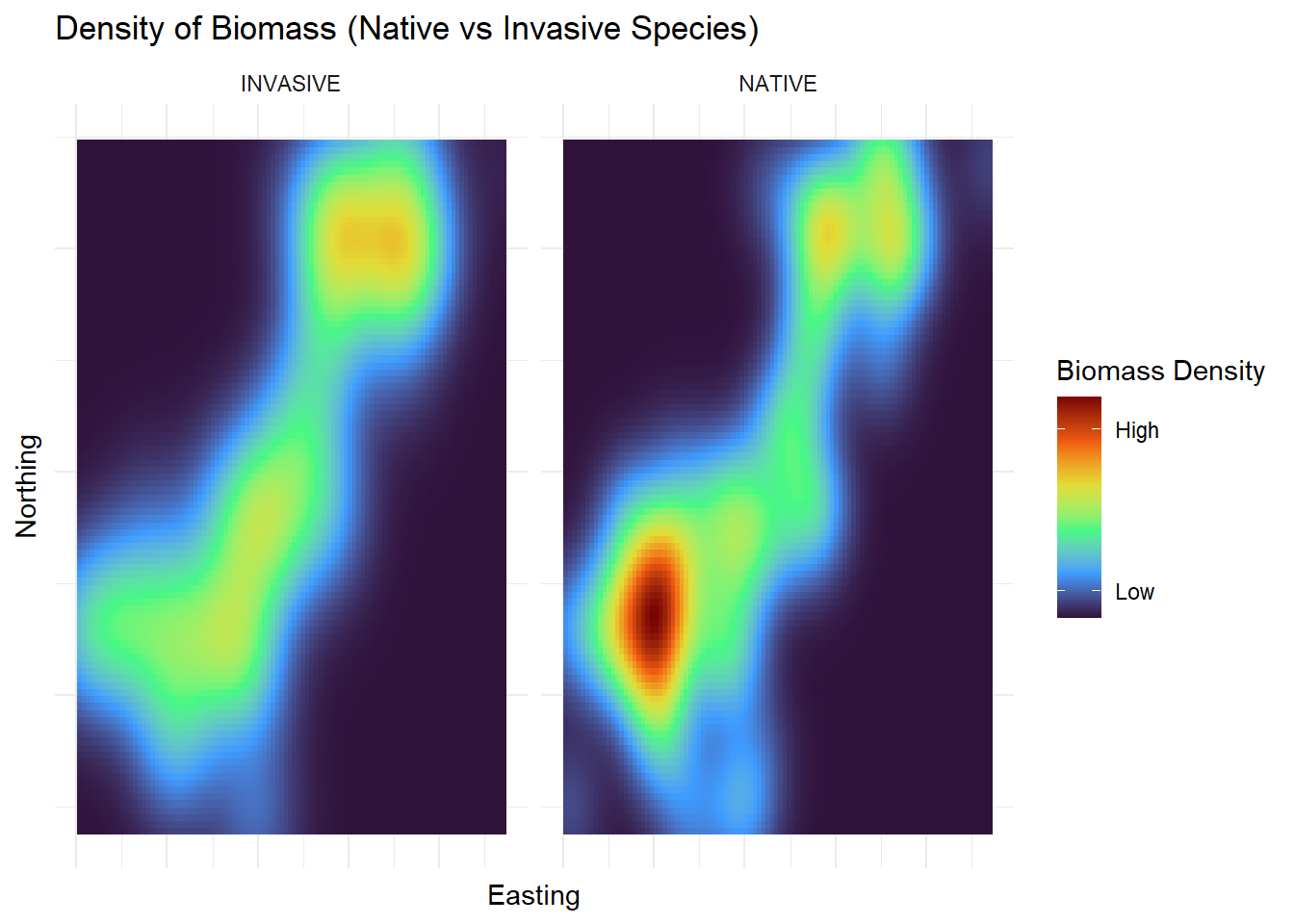
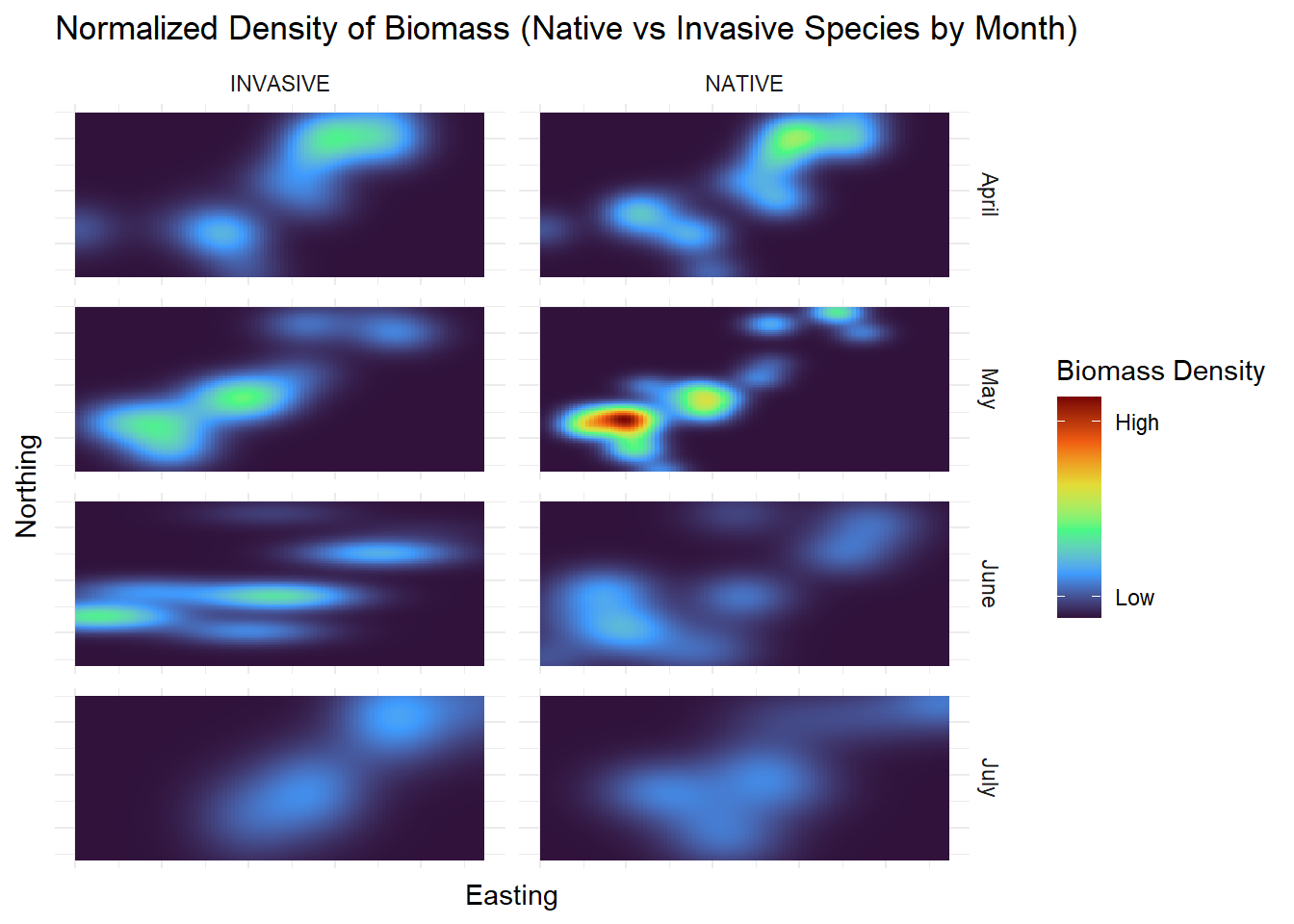
Categorical |
Each species is categorized as native or invasive |
| Season |
Categorical |
Our observations are grouped based on the date that they were sampled (Spring, Summer and Fall) to observe the changes in nutritional quality |
| Dates |
Categorical |
This keeps track of the day that each observation was sampled |
| Aspect |
Categorical |
Described the direction the hill was facing that each of our transects had been sampled on |
| Elev |
Quantitative |
Describes the elevation that each of the transects was sampled at |
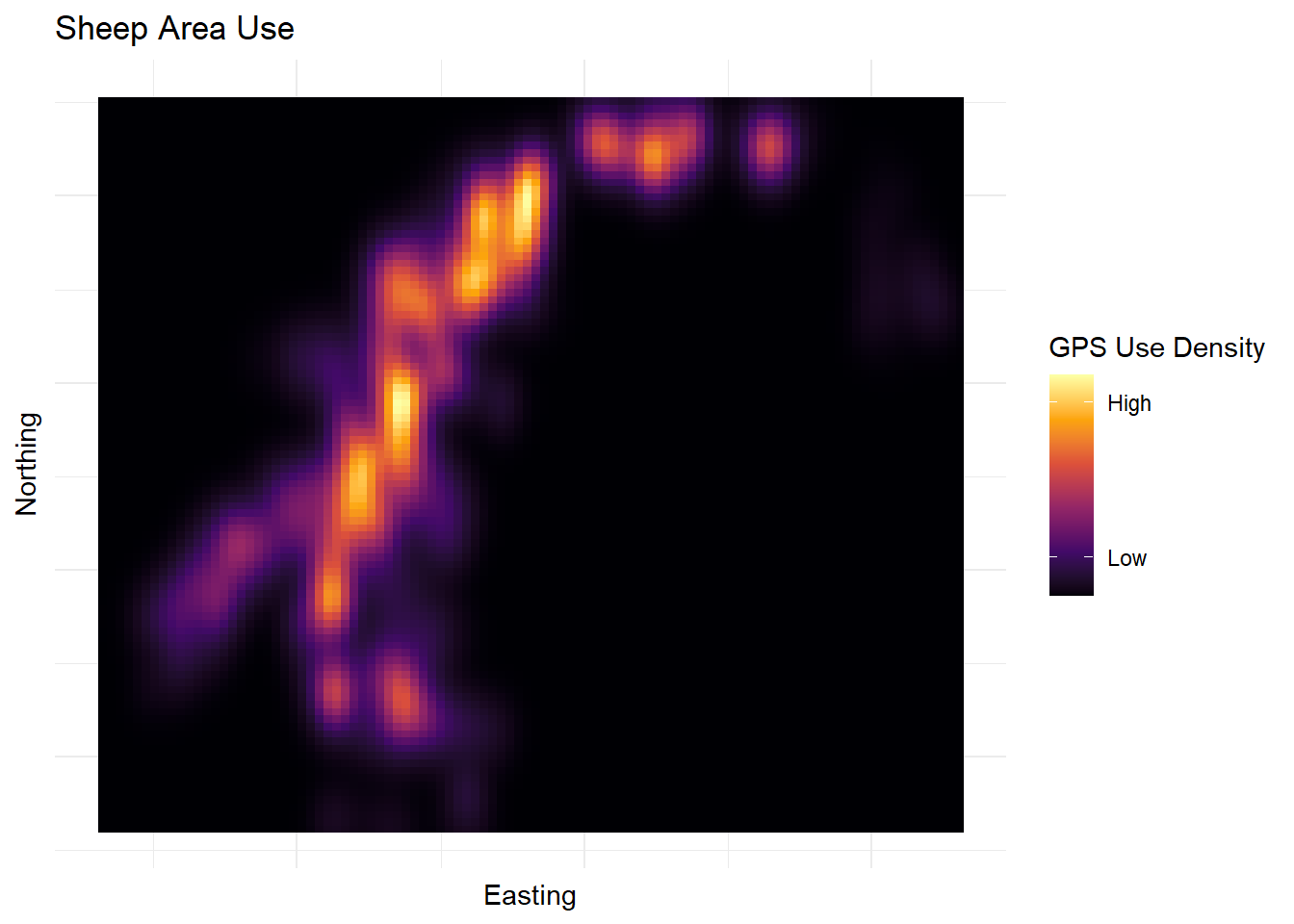
| Lat/Long |
Categorical |
Each of the lat/long pairs plots the beginning, middle and end of each of the transects |
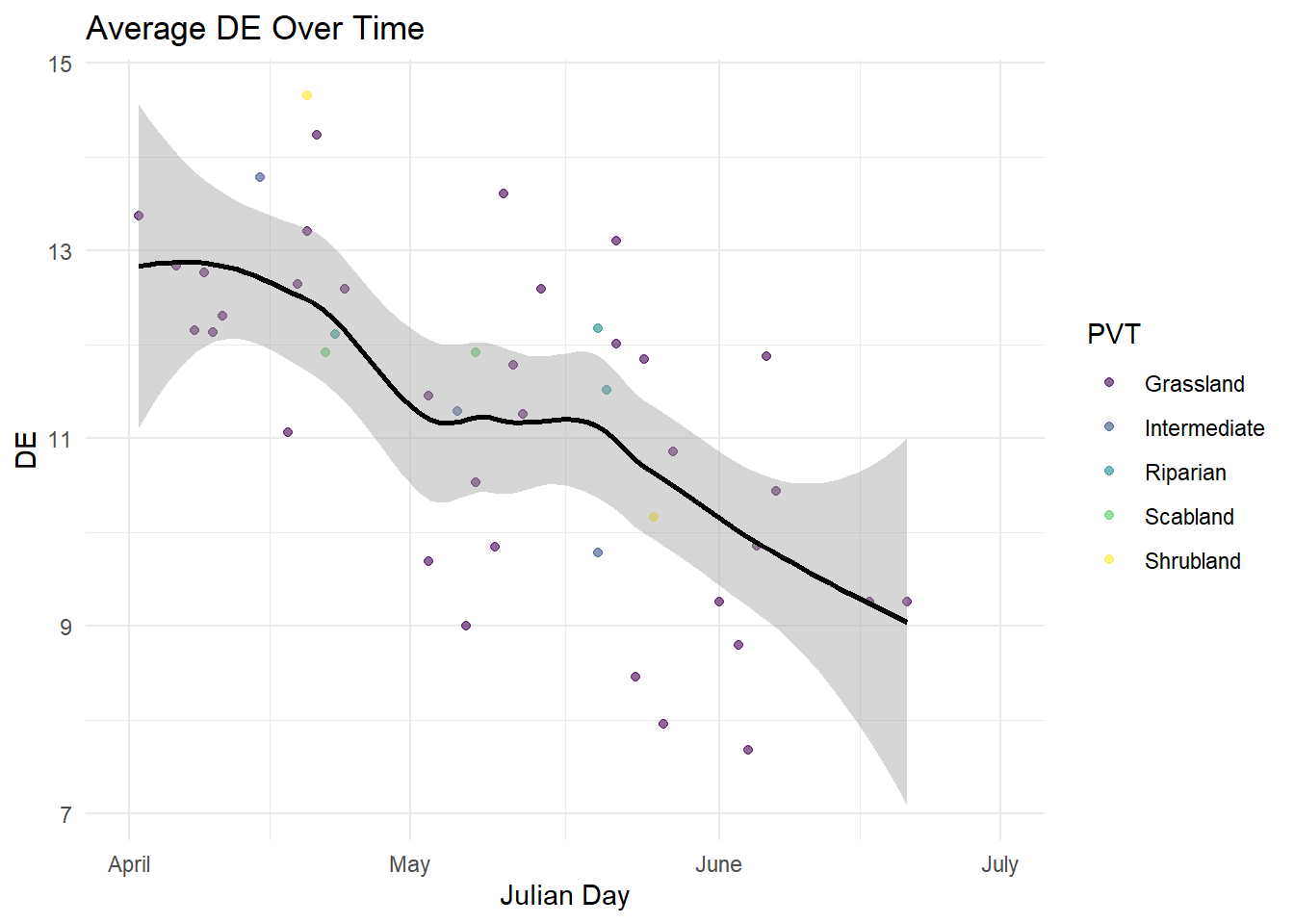
| DE |
Quantitative |
Amount of digestible energy provided by each species |
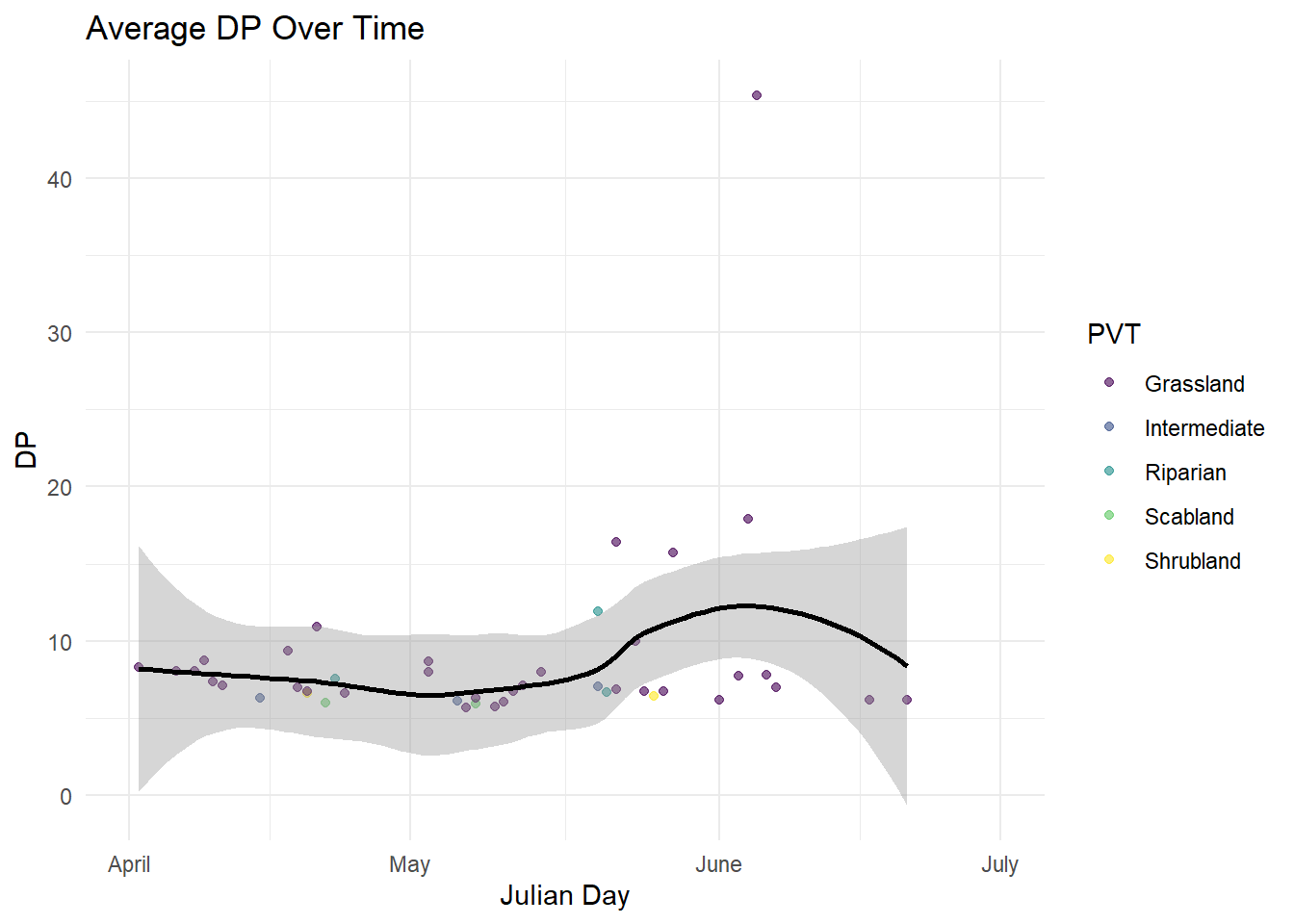
| DP |
Quantitative |
Amount of digestible protien provided by each species |
| DE.SD |
Quantitative |
Standard deviation of digestible energy |
| DP.SD |
Quantitative |
Standard deviation of digestible protien |
| Julian Date |
Categorical |
The converted date that each of the plots was sampled to julian day |
| Easting |
Categorical |
The converted latitude to UTM |
| Northing |
Categorical |
The converted longitude to UTM |
| TotalDE |
Quantitative |
The amount of total DE available at a transect |
| TotalDP |
Quantitative |
The amount of total DP available at a transect |
| SuitableBiomass |
Quantitative |
The amount of forage available at each transect that meet the assigned energetic demands of a lactating female sheep |
| AveDE |
Quantitative |
The average amount of digestible energy available at a transect |
| AveDP |
Quantitative |
The average amount of digestible protien available at a transect |
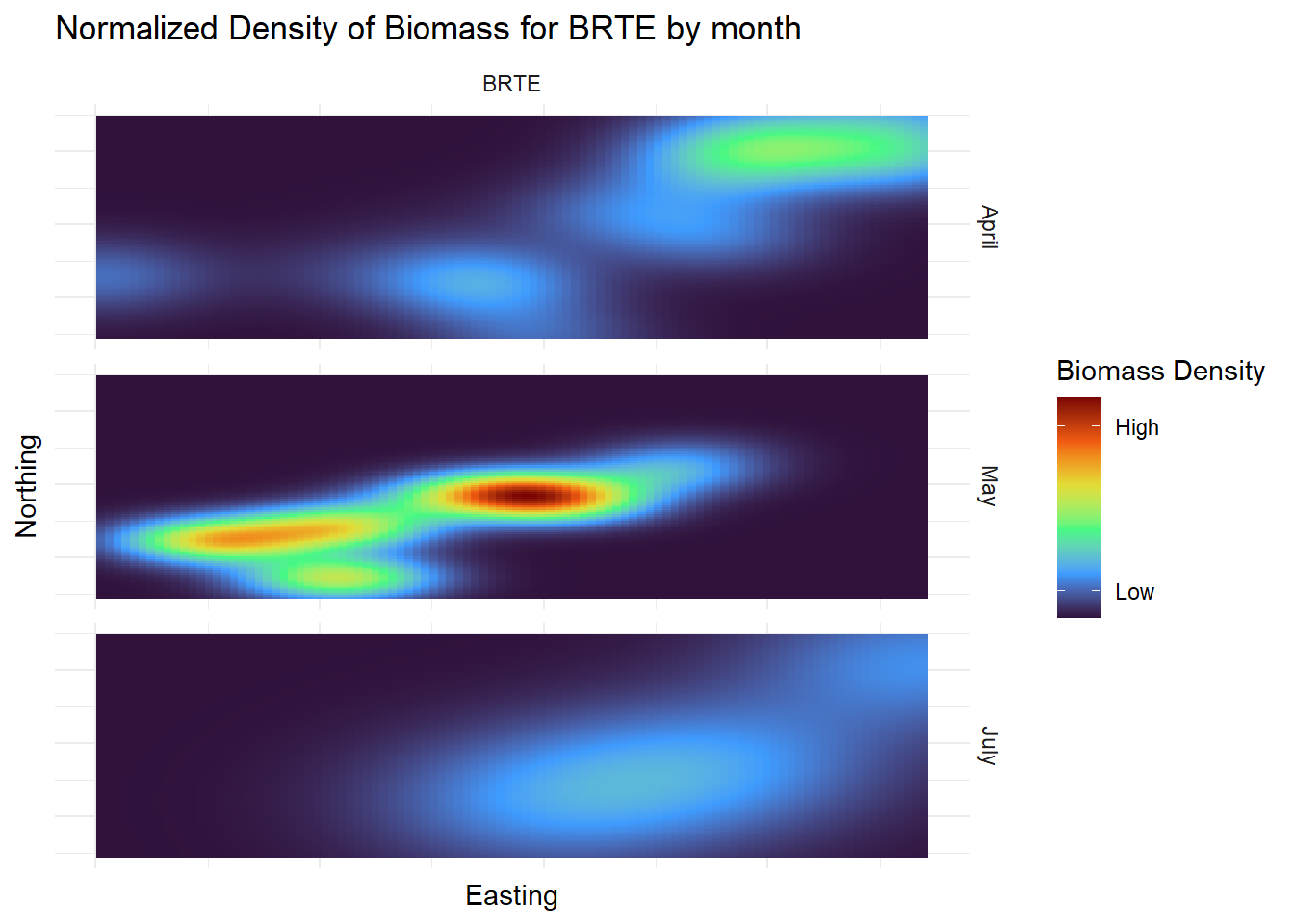
| Month |
Categorical |
The month that the plot was sampled |